Making Beautiful Tables with R
Use groups instead of repetitive columns
This lesson is called Use groups instead of repetitive columns, part of the Making Beautiful Tables with R course. This lesson is called Use groups instead of repetitive columns, part of the Making Beautiful Tables with R course.
Transcript
Click on the transcript to go to that point in the video. Please note that transcripts are auto generated and may contain minor inaccuracies.
It’s better to use groups instead of repetitive columns. For instance, the island column repeats itself multiple times before we move on to the next island name. This is a doesn’t really look good. And it would be nice if we can just make it visually clearer that certain rows belong to certain islands.
The way to do that is to use groups. This means that we have rows (one for each island) that spans all of the columns. Here, we’ll transition to as_flextable() because I like hide_grouplabel. But I’ll show the flextable() version before that.
penguin_counts_wider |>
mutate(island = paste('Island: ', island)) |>
as_grouped_data(
groups = 'island'
) |>
as_flextable(hide_grouplabel = TRUE) |>
set_header_labels(
island = 'Island',
year = '',
Adelie_female = 'Female',
Adelie_male = 'Male',
Chinstrap_female = 'Female',
Chinstrap_male = 'Male',
Gentoo_female = 'Female',
Gentoo_male = 'Male'
) |>
add_header_row(
values = c('', 'Adelie', 'Chinstrap', 'Gentoo'),
colwidths = c(1, 2, 2, 2)
) |>
add_header_lines(
values = c('Penguins in the Palmer Archipelago', 'Data is courtesy of the {palmerpenguins} R package')
) |>
align(i = 3, align = 'center', part = 'header') |>
autofit()Your Turn
Group your table by continent. Using hide_grouplabel = TRUE in as_flextable() may be helpful. At the end, your table should look like this:
Have any questions? Put them below and we will help you out!
Course Content
16 Lessons
You need to be signed-in to comment on this post. Login.
Ahmad Rabiee • October 11, 2023
Hi When I run these codes, I get this error message (see below). Can you please help, not sure what I am doing wrong here? Ahmad
Albert Rapp • October 12, 2023
Hi Ahmad,
when you switch to groups you actually "lose" one column. That's why in
add_header_rows()you will need to modifycolwidths= c(2,2,2,2)tocolwidths= c(1,2,2,2). It's easy to forget this change. That's what happened to me in the video at 2:22.Best, Albert
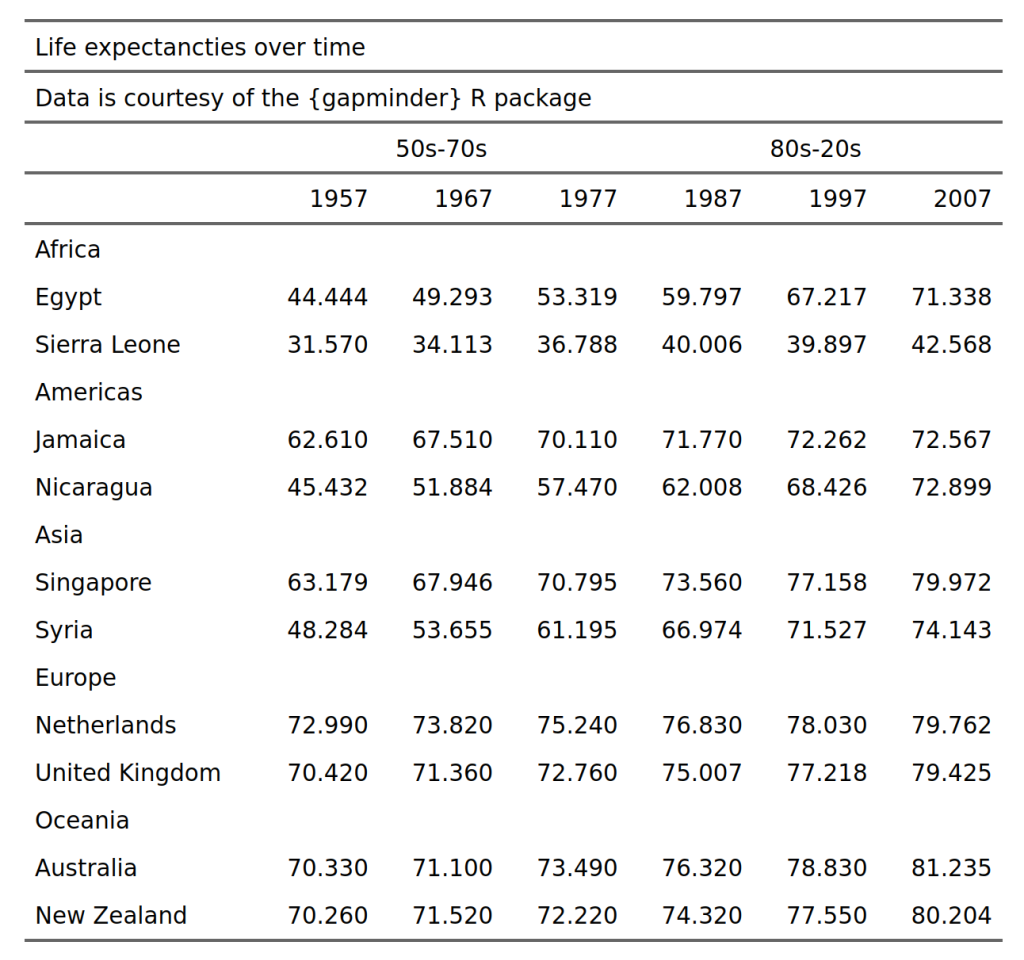
Lorenzo Botto • June 29, 2024
Hi, In the table above from gapminder, I would like to keep the continent left aligned but each country within each continent right aligned. When I use the line
align(j = 'country', part = 'body', align = 'right')both the continent and the countries are aligned to the right. Thank you, LorenzoAlbert Rapp • July 1, 2024
Due to the grouping, the things that are currently in the continent column are treated as if they were extra rows in the country column when the table is assembled. That's why the continents get right-aligned too. In this case, you will have to specify which rows to right-align via the
iargument inalign(). The best way to do that is to specify that the rows where the column country in the data set are NAs. You can see how the grouped data set looks by executing onlyAnd to target the desired rows you will have to use the tilde notation (which we will use extensively when learning about styling). The code for this looks like this: